To provide a comprehensive knowledgebase for curation of human proteins with differential expression during the HIV infection, replication and latency, this protein-centric database collected experimental studies of differentially regulated genes. Then the extracted genes were further mapped to their protein products, with detailed protein functional and structural annotations presented. All the curated datasets are publicly available. This database covers a wide range of experimental studies and bridges the gap between our understanding of differentially expressed human genes, protein products, and functional annotations during the HIV infection, replication and latency. In particular, the quantitative data extracted from the experimental studies is made available and displayed in the “Gene Expression Profile” tab for each entry in our database.
We believe HIVed is of great benefit and has important implications for HIV studies due to its ease of use and ability to display common threads among various HIV latency and infection datasets. In particular, this database allows users to compare the gene expression studies during HIV latency for their genes of interest. Users, for example, can easily identify proteins that are up- or down-regulated across all the HIV infection, replication and latency studies, and verify if a particular protein has been identified as an HIV replication factor and if it is a druggable target. This is very useful for identifying novel protein targets for combating the HIV. Additionally, this database has the ability to update entries resulting from up-to-date studies pertinent to HIV latency, replication, and infection, enabling it to keep pace with the rapid proliferation of HIV studies.
Notice
1. In this database, some genes could not be mapped to the UniProt database while some genes were mapped to multiple UniProt database. The lists for those genes can be found in the Help webpage (sections 2 and 3). When searching protein products for those genes that were mapped to multiple UniProt IDs, please bear in mind that such genes with different UniProt IDs will have the same gene expression profile as we used the gene name to extract the gene expression data from the sutdies we curated in this work.
2. We have noticed that some genes were annotated as homologous genes of homo sapiens according to the original data files from the papers. As such, these genes were annotated to be from other species. We have accordingly removed the expression profile of these genes from our database. However, there still could be some genes that have been missed. Please let us know if you find anything wrong with the gene expression data and we will manually check and update the database accordingly.
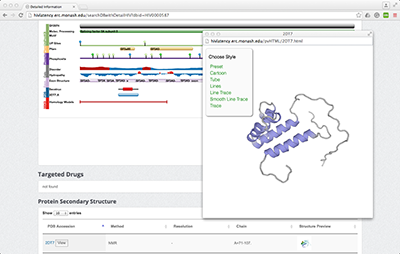
3. The protein feature viewer plugin is unavailable anymore due to the recent significant update of PDB database.
Citing us
If you found our HIVed database useful, please consider citing us: Chen Li, Sri H Ramarathinam, Jerico Revote, Georges Khoury, Jiangning Song, Anthony W Purcell. HIVed, a knowledgebase for differentially expressed human genes and proteins during HIV infection, replication and latency. Scientific Reports, 7, 45509, 2017.